Validation of Recombinant Human Proteins with High-Resolution Mass Spectrometry
Introduction
In May 2010, OriGene and the Institute for Systems Biology (ISB) announced a collaboration to create a SRM/MRM Mass Spectrometry (MS) Standard Database for 5,000 Human Proteins. Learn more

As the first step of this project, OriGene has delivered recombinant purified human proteins to ISB for mass spec validation. These proteins are made in mammalian cells with authentic protein structures. Learn more
Workflow
Scientists at ISB apply these protein samples to mass spectrometer to obtain high quality proteotypic peptide sequences, mapping, and display through the PeptideAtlas database as outlined briefly below:
A highly purified OriGene protein sample is digested with trypsin.
- The sample is run through high-resolution Ion-Trap and Quadrupole-Tof mass spectrometers (e.g., ESI MS/MS, ETD etc).
- The MS/MS spectra are compared to theoretical spectra (SEQUEST, X!Tandem) or actual spectra (SpectraST) to identify all possible peptides and their post-translational modifications.
- The peptide identifications are scored, formed into false and true positive distributions, and subsequently filtered to retain only the highest scoring identifications (PeptideProphet).
- The peptide sequences are compared to protein sequence databases (e.g. for human, we use the Ensembl, IPI, and Swiss-Prot protein sequence databases). As the peptides are identified in a given protein, so are their locations relative to the protein start (CDS coordinates).
- The peptide locations in chromosomal coordinates are calculated from the CDS coordinates.
- The protein identifications are clustered and annotated (ProteinProphet) and stored in the publically accessible PeptideAtlas.
Typical Results
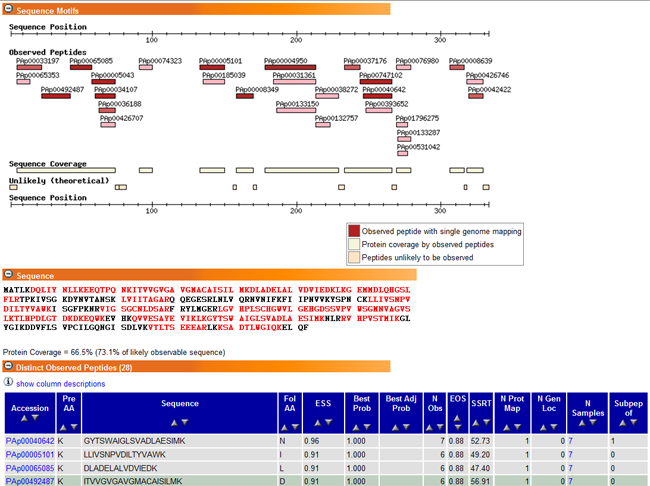
So far the Moritz group at ISB has completed MS/MS analysis for over 3000 proteins provided by OriGene and has positively confirmed these proteins as expected from the expression constructs. Below is a screen shot of selected data on LDHA.
Goals
The long term goal of this collaboration is to develop a human Peptide- and SRM-Atlas to provide MRM/SRM MS assays for all 5,000 OriGene human expressed proteins.
Learn more about OriGene’s heavy isotope labeled proteins for MRM/SRM MS application.
View examples of SILAC lablelled human expressed MAPK1 and MAP2K1 OriGene proteins.
Contact us if you are interested in making custom heavy labeled proteins or peptides.
Recombinant Protein Resources |






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China