Glutathione Peroxidase 4 (GPX4) Human Gene Knockout Kit (CRISPR)
CAT#: KN408065
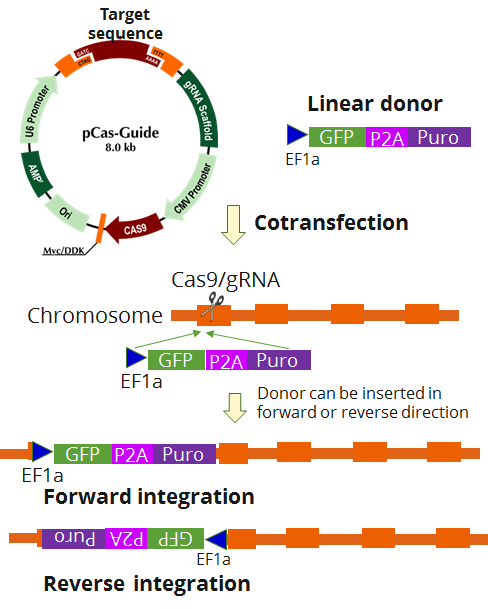
GPX4 - KN2.0, Human gene knockout kit via CRISPR, non-homology mediated.
KN2.0 knockout kit validation
USD 1,657.00
2 Weeks*
USD 450.00
USD 300.00
Specifications
| Product Data | |
| Format | 2 gRNA vectors, 1 linear donor |
| Donor DNA | EF1a-GFP-P2A-Puro |
| Symbol | Glutathione Peroxidase 4 |
| Locus ID | 2879 |
| Components |
KN408065G1, Glutathione Peroxidase 4 gRNA vector 1 in pCas-Guide CRISPR vector, Target Sequence: GCTTCAGTAGGCGGCAAAGG KN408065G2, Glutathione Peroxidase 4 gRNA vector 2 in pCas-Guide CRISPR vector, Target Sequence: GGCGCTGCTCTGTGGGGCTC KN408065D, Linear donor DNA containing LoxP-EF1A-tGFP-P2A-Puro-LoxP: The sequence below is cassette sequence only. The linear donor DNA also contains proprietary target sequence. LoxP-EF1A-tGFP-P2A-Puro-LoxP (2739 bp) ATAACTTCGT ATAATGTATG CTATACGAAG TTATCGTGAG GCTCCGGTGC CCGTCAGTGG GCAGAGCGCA CATCGCCCAC AGTCCCCGAG AAGTTGGGGG GAGGGGTCGG CAATTGAACC GGTGCCTAGA GAAGGTGGCG CGGGGTAAAC TGGGAAAGTG ATGTCGTGTA CTGGCTCCGC CTTTTTCCCG AGGGTGGGGG AGAACCGTAT ATAAGTGCAG TAGTCGCCGT GAACGTTCTT TTTCGCAACG GGTTTGCCGC CAGAACACAG GTAAGTGCCG TGTGTGGTTC CCGCGGGCCT GGCCTCTTTA CGGGTTATGG CCCTTGCGTG CCTTGAATTA CTTCCACCTG GCTGCAGTAC GTGATTCTTG ATCCCGAGCT TCGGGTTGGA AGTGGGTGGG AGAGTTCGAG GCCTTGCGCT TAAGGAGCCC CTTCGCCTCG TGCTTGAGTT GAGGCCTGGC CTGGGCGCTG GGGCCGCCGC GTGCGAATCT GGTGGCACCT TCGCGCCTGT CTCGCTGCTT TCGATAAGTC TCTAGCCATT TAAAATTTTT GATGACCTGC TGCGACGCTT TTTTTCTGGC AAGATAGTCT TGTAAATGCG GGCCAAGATC TGCACACTGG TATTTCGGTT TTTGGGGCCG CGGGCGGCGA CGGGGCCCGT GCGTCCCAGC GCACATGTTC GGCGAGGCGG GGCCTGCGAG CGCGGCCACC GAGAATCGGA CGGGGGTAGT CTCAAGCTGG CCGGCCTGCT CTGGTGCCTG GCCTCGCGCC GCCGTGTATC GCCCCGCCCT GGGCGGCAAG GCTGGCCCGG TCGGCACCAG TTGCGTGAGC GGAAAGATGG CCGCTTCCCG GCCCTGCTGC AGGGAGCTCA AAATGGAGGA CGCGGCGCTC GGGAGAGCGG GCGGGTGAGT CACCCACACA AAGGAAAAGG GCCTTTCCGT CCTCAGCCGT CGCTTCATGT GACTCCACGG AGTACCGGGC GCCGTCCAGG CACCTCGATT AGTTCTCGAG CTTTTGGAGT ACGTCGTCTT TAGGTTGGGG GGAGGGGTTT TATGCGATGG AGTTTCCCCA CACTGAGTGG GTGGAGACTG AAGTTAGGCC AGCTTGGCAC TTGATGTAAT TCTCCTTGGA ATTTGCCCTT TTTGAGTTTG GATCTTGGTT CATTCTCAAG CCTCAGACAG TGGTTCAAAG TTTTTTTCTT CCATTTCAGG TGTCGTGAAT GGAGAGCGAC GAGAGCGGCC TGCCCGCCAT GGAGATCGAG TGCCGCATCA CCGGCACCCT GAACGGCGTG GAGTTCGAGC TGGTGGGCGG CGGAGAGGGC ACCCCCGAGC AGGGCCGCAT GACCAACAAG ATGAAGAGCA CCAAAGGCGC CCTGACCTTC AGCCCCTACC TGCTGAGCCA CGTGATGGGC TACGGCTTCT ACCACTTCGG CACCTACCCC AGCGGCTACG AGAACCCCTT CCTGCACGCC ATCAACAACG GCGGCTACAC CAACACCCGC ATCGAGAAGT ACGAGGACGG CGGCGTGCTG CACGTGAGCT TCAGCTACCG CTACGAGGCC GGCCGCGTGA TCGGCGACTT CAAGGTGATG GGCACCGGCT TCCCCGAGGA CAGCGTGATC TTCACCGACA AGATCATCCG CAGCAACGCC ACCGTGGAGC ACCTGCACCC CATGGGCGAT AACGATCTGG ATGGCAGCTT CACCCGCACC TTCAGCCTGC GCGACGGCGG CTACTACAGC TCCGTGGTGG ACAGCCACAT GCACTTCAAG AGCGCCATCC ACCCCAGCAT CCTGCAGAAC GGGGGCCCCA TGTTCGCCTT CCGCCGCGTG GAGGAGGATC ACAGCAACAC CGAGCTGGGC ATCGTGGAGT ACCAGCACGC CTTCAAGACC CCGGATGCAG ATGCCGGTGA AGAAAGAGGA AGCGGAGCTA CTAACTTCAG CCTGCTGAAG CAGGCTGGAG ACGTGGAGGA GAACCCTGGA CCTATGACCG AGTACAAGCC CACGGTGCGC CTCGCCACCC GCGACGACGT CCCCAGGGCC GTACGCACCC TCGCCGCCGC GTTCGCCGAC TACCCCGCCA CGCGCCACAC CGTCGATCCG GACCGCCACA TCGAGCGGGT CACCGAGCTG CAAGAACTCT TCCTCACGCG CGTCGGGCTC GACATCGGCA AGGTGTGGGT CGCGGACGAC GGCGCCGCGG TGGCGGTCTG GACCACGCCG GAGAGCGTCG AAGCGGGGGC GGTGTTCGCC GAGATCGGCC CGCGCATGGC CGAGTTGAGC GGTTCCCGGC TGGCCGCGCA GCAACAGATG GAAGGCCTCC TGGCGCCGCA CCGGCCCAAG GAGCCCGCGT GGTTCCTGGC CACCGTCGGC GTCTCGCCCG ACCACCAGGG CAAGGGTCTG GGCAGCGCCG TCGTGCTCCC CGGAGTGGAG GCGGCCGAGC GCGCCGGGGT GCCCGCCTTC CTGGAGACCT CCGCGCCCCG CAACCTCCCC TTCTACGAGC GGCTCGGCTT CACCGTCACC GCCGACGTCG AGGTGCCCGA AGGACCGCGC ACCTGGTGCA TGACCCGCAA GCCCGGTGCC TGAAACTTGT TTATTGCAGC TTATAATGGT TACAAATAAA GCAATAGCAT CACAAATTTC ACAAATAAAG CATTTTTTTC ACTGCATTCT AGTTGTGGTT TGTCCAAACT CATCAATGTA TCTTAATAAC TTCGTATAAT GTATGCTATA CGAAGTTAT
|
| Disclaimer | These products are manufactured and supplied by OriGene under license from ERS. The kit is designed based on the best knowledge of CRISPR technology. The system has been functionally validated for knocking-in the cassette downstream the native promoter. The efficiency of the knock-out varies due to the nature of the biology and the complexity of the experimental process. |
| Reference Data | |
| RefSeq | NM_001039847, NM_001039848, NM_002085, NM_001367832 |
| UniProt ID | P36969 |
| Synonyms | GPx-4; GSHPx-4; MCSP; PHGPx; snGPx; snPHGPx |
| Summary | The protein encoded by this gene belongs to the glutathione peroxidase family, members of which catalyze the reduction of hydrogen peroxide, organic hydroperoxides and lipid hydroperoxides, and thereby protect cells against oxidative damage. Several isozymes of this gene family exist in vertebrates, which vary in cellular location and substrate specificity. This isozyme has a high preference for lipid hydroperoxides and protects cells against membrane lipid peroxidation and cell death. It is also required for normal sperm development; thus, it has been identified as a 'moonlighting' protein because of its ability to serve dual functions as a peroxidase, as well as a structural protein in mature spermatozoa. Mutations in this gene are associated with Sedaghatian type of spondylometaphyseal dysplasia (SMDS). This isozyme is also a selenoprotein, containing the rare amino acid selenocysteine (Sec) at its active site. Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. Transcript variants resulting from alternative splicing or use of alternate promoters have been described to encode isoforms with different subcellular localization. [provided by RefSeq, Dec 2018] |
Documents
| Product Manuals |
| FAQs |
| SDS |
Resources
Other Versions
| SKU | Description | Size | Price |
|---|---|---|---|
| GA101938 | GPX4 CRISPRa kit - CRISPR gene activation of human glutathione peroxidase 4 |
USD 1,657.00 |
{0} Product Review(s)
Be the first one to submit a review






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China