ASOs vs siRNA – Which is Right for Me?
Dec 21, 2023

Antisense Oligonucleotides (ASOs) and small interfering RNAs (siRNAs) emerge as pivotal tools in molecular biology, offering precise means to silence targeted genes. In this blog we will explore the intricacies of these powerful gene-silencing tools, delving into their mechanisms and key differences.
What are Antisense Oligonucleotides?
Antisense oligonucleotides (ASOs) are synthetic, single-stranded DNA or RNA molecules designed to bind selectively to complementary RNA sequences. On average, they range from 15-25 nucleotides (nt) in length1. ASOs work by modulating the expression of specific genes by targeting messenger RNA (mRNA). The concept of ASOs relies on the ability of these synthetic molecules to hybridize with target RNA sequences, thereby influencing gene expression through various mechanisms.
The primary mechanism involves the formation of RNA-DNA heteroduplexes between the ASO and the complementary mRNA sequence. This binding triggers the recruitment of the enzyme RNase H, which specifically cleaves the RNA strand within the hybrid, leading to the degradation of the targeted mRNA. This degradation impedes the translation process, resulting in reduced protein expression from the targeted gene.
Additionally, ASOs can interfere with mRNA maturation by hindering the crucial 5' cap formation. Targeting specific mRNA regions, ASOs disrupt the addition of the 5' cap, destabilizing the mRNA and diminishing its translation efficiency. ASOs also possess the capability to influence pre-mRNA splicing events, a mechanism known as splice-switching. By hybridizing with pre-mRNA sequences, ASOs can alter splice site recognition, leading to the inclusion or exclusion of exons during mRNA processing. This modification in splicing patterns results in the production of altered mRNA isoforms or the exclusion of targeted exons, affecting the resulting protein products or triggering mRNA degradation.
Furthermore, ASOs can physically obstruct ribosomal activity by binding to mRNA sequences near translation initiation sites or along coding regions. This steric hindrance impedes ribosome binding or progression, ultimately reducing protein synthesis efficiency2.
What are siRNAs?
Small interfering RNA (siRNA) refers to short double-stranded RNA molecules typically about 20-25 nucleotides in length. siRNA plays a crucial role in the regulation of gene expression through a process called RNA interference (RNAi).
This process commences with the generation of siRNA molecules, either by introducing double-stranded RNA (dsRNA) or via enzymatic cleavage of long dsRNA by the enzyme, Dicer. The produced siRNAs are then loaded onto the RNA-induced silencing complex (RISC), where one strand acts as the guide. This guide strand selectively binds to complementary mRNA sequences, guiding the RISC complex to the targeted mRNA. Upon binding, siRNA directs the cleavage of the mRNA by the RISC complex, resulting in mRNA degradation and halting translation. RNAi, driven by siRNA, enables precise post-transcriptional gene silencing, playing pivotal roles in gene expression regulation, cellular processes, and offering researchers a potent tool for investigating gene function and potential therapeutic interventions. A detailed graphic of this process can be seen below:
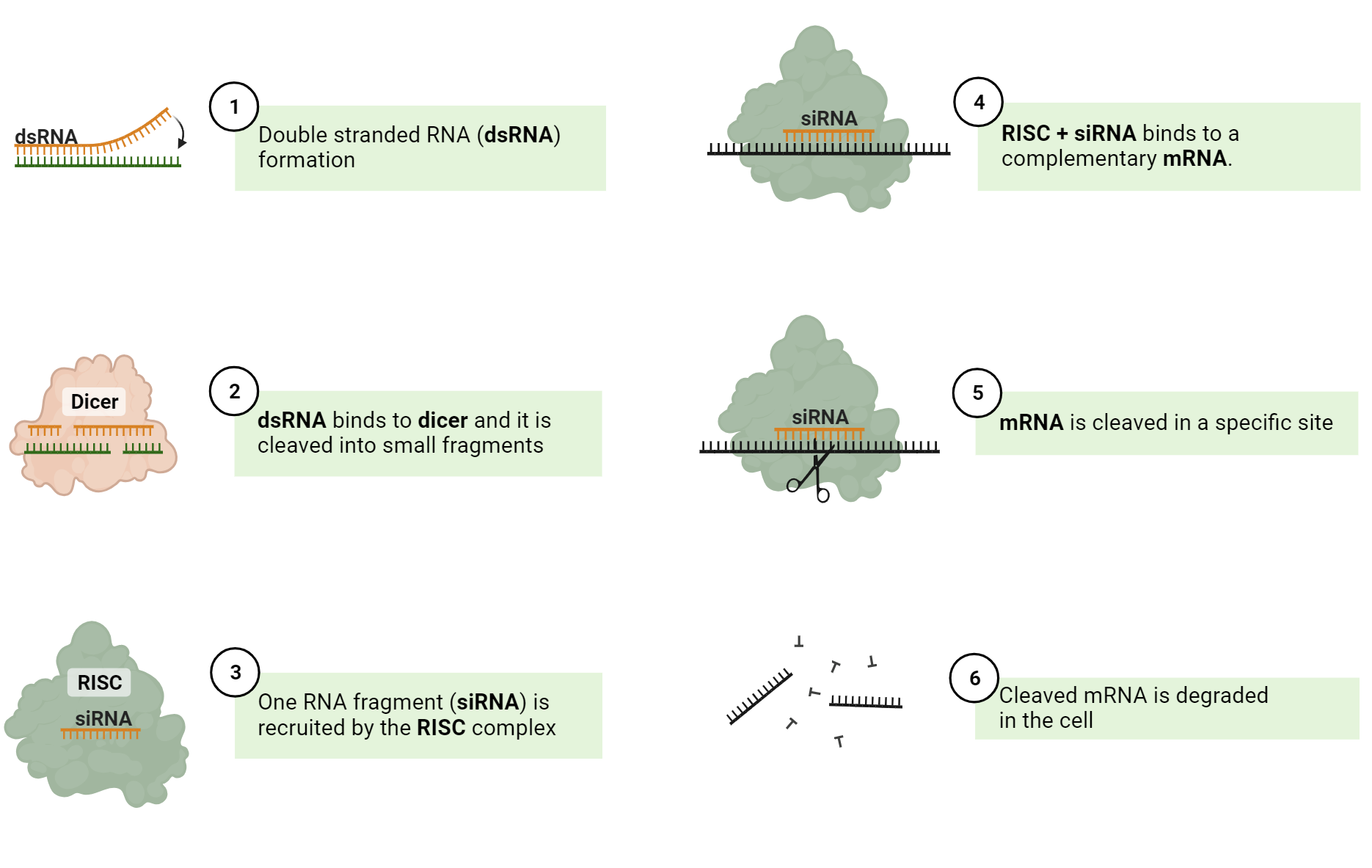
Key Differences between ASOs and siRNA
Origin and Structure:
ASOs: ASOs are synthetic, single-stranded DNA or RNA molecules designed to be complementary to the target mRNA sequence.
siRNA: siRNA molecules are typically double-stranded RNA molecules, about 20-25 nucleotides in length, generated from longer dsRNA or through enzymatic processing.
Mechanism of Action:
ASOs: ASOs modulate gene expression by directly binding to complementary mRNA sequences. ASOs can inhibit mRNA translation by blocking ribosome binding or induce mRNA degradation via RNase H-mediated cleavage3.
siRNA: siRNA operates through RNA interference (RNAi). Once loaded into the RNA-induced silencing complex (RISC), siRNA guides the RISC to complementary mRNA, leading to mRNA cleavage and degradation, preventing translation.
Delivery Challenges:
ASOs: ASOs also face delivery challenges, but their single-stranded nature might offer certain advantages in terms of stability and delivery compared to siRNA.
siRNA: Delivery of siRNA can be challenging due to its susceptibility to degradation and issues related to cellular uptake.
Cost Considerations:
ASOs: ASOs, being single-stranded and synthetically designed, might offer potential cost advantages in production compared to siRNA due to simpler synthesis methods and purification.
siRNA: Production of double-stranded siRNA might involve higher manufacturing costs due to synthesis complexity and purification methods.
Considerations When Choosing Between ASOs and siRNAs
When choosing between antisense oligonucleotides (ASOs) and small interfering RNAs (siRNAs) for cell culture experiments, siRNAs often emerge as the preferred option due to their relative simplicity in discovering potent molecules and ease of obtaining unmodified RNA with high potency. ASOs, conversely, require chemical modifications to ensure activity inside cells3. However, the decision becomes more intricate when developing compounds for animal testing or therapeutic investigations.
The unresolved choice between ASOs and siRNAs in vivo will likely evolve over the coming years, considering the challenges of delivering oligonucleotides to specific target tissues. Both platforms share similarities as gene expression modulators, yet differ in the number of strands they possess, potentially impacting cost and delivery complexity.
Optimizing both technologies typically yield potent and effective molecules, as an ideal target sequence for an ASO might not align with an ideal sequence for an siRNA, and vice versa. SiRNAs have exhibited robustness in cell culture, but their superiority in vivo remains uncertain, driving ongoing advancements in their development4.
Ultimately, the choice between ASOs and siRNAs hinges on a comprehensive evaluation of an individual’s experimental objectives, accessibility of technology, and delivery considerations.
RNAi Solutions for Researchers
OriGene offers a spectrum of gene silencing solutions, including siRNA oligo duplexes, shRNA kits, and miRNA plasmids. Uncover the depth and versatility of OriGene's offerings as we pave the way for transformative discoveries in gene modulation and molecular research.
Citation
- Sohail M, Hochegger H, Klotzbücher A, Guellec RL, Hunt T, Southern EM. Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res. 2001 May 15;29(10):2041-51. doi: 10.1093/nar/29.10.2041. PMID: 11353073; PMCID: PMC55457.
- Di Fusco D, Dinallo V, Marafini I, Figliuzzi MM, Romano B, Monteleone G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front Pharmacol. 2019 Mar 29;10:305. doi: 10.3389/fphar.2019.00305. PMID: 30983999; PMCID: PMC6450224.
- Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2012 Jan;226(2):365-79. doi: 10.1002/path.2993. Epub 2011 Nov 9. PMID: 22069063; PMCID: PMC3916955.
Infographics generated with Biorender






























































































































































































































































 Germany
Germany
 Japan
Japan
 United Kingdom
United Kingdom
 China
China